
Laboratory for
Epigenome Inheritance
RIKEN IMS, Yokohama, Japan
Epigenetic Inheritance from Oocytes
Parental traits are transmitted to the offspring via DNA. Gametes (oocyte and sperm) are responsible for this transmission. They contain not only DNA but also epigenetic information (epigenome) which is the instruction manual for decoding DNA functions. Thanks to the epigenetic inheritance across generations, the next generation embryos can develop properly. Our laboratory studies how the epigenome is established during gametogenesis in the parental generation, how it is passed on to the next generation, and how it works. Furthermore, we examine whether environmental factors in the parental generation affect the intergenerational epigenetic inheritance, which can be viewed as an attempt to reexamine the Lamarckism (inheritance of acquired traits) using the latest scientific technology. For this purpose, we are studying (1) the mechanism of epigenome establishment during gametogenesis and its transmission to the next generation, (2) the significance of epigenetic inheritance, and (3) the mechanism of disease predisposition formation via inheritable epigenome, mainly using mouse as a model. These studies provide a molecular foundation for understanding how early molecular events during gametogenesis and early embryogenesis impact on later development and potentially on phenotypes in the offspring.
Keywords: Histone, Reprogramming, Oocyte, Preimplantation embryo
Current research directions
1. Understanding mechanisms of epigenome establishment in oocytes
2. Understanding functions and mechanisms of epigenetic inheritance and reprogramming
3. Understanding the effects of maternal environment and diseases on epigenetic inheritance
Preimplantation embryos
A new life begins when a sperm enters an oocyte. The two originally different cells are now fused to form a single cell, called zygote, in which two nuclei co-exist (paternal pronucleus and maternal pronucleus). The zygote divides to form a 2-cell embryo and the subsequent cleavages give rise to 4-cell, morula (8-16 cells), and blastocyst (32-128 cells) embryos. This developmental process takes place in the oviduct and the uterus before implantation and therefore is called ‘pre-implantation development’. Preimplantation embryos can be cultured in vitro up to the blastocyst stage, and they can give rise to live animals after transplanted into surrogate mothers. Such 'ex vivo' permissive characteristics of preimplantation embryos allow researchers to easily manipulate gene function in a physiological context. Importantly, the epigenetic landscape is genome-widely reprogrammed in preimplantation embryos, providing an excellent model to study epigenetics.


Epigenetics
The epigenetics field is getting bigger and bigger. Epigenetics is nowadays an infrastructure to study life sciences. There are various definitions of epigenetics, but my understanding of epigenetics is a gene regulatory mechanism beyond the layers of genomic sequence and transcription factors. The nature of epigenetics include chemical modifications on DNA and histones. Epigenetic modifications compartmentalizes chromatin into transcriptionally-permissive and -repressive states. Each cell type has its own epigenetic landscape to enable a cell-type specific gene expression program. See more information about Epigenetics elsewhere if needed.
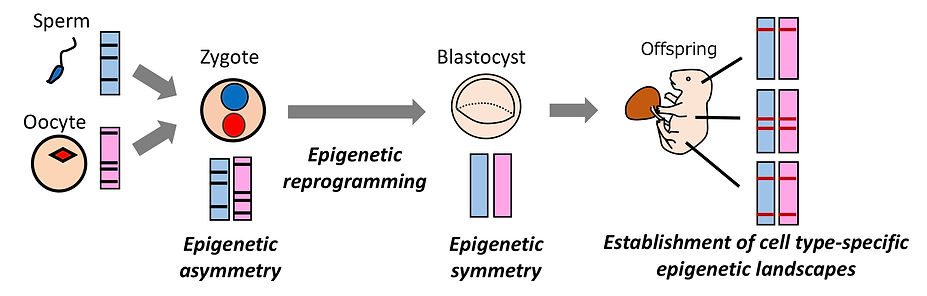
Epigenetic reprogramming
The epigenetic landscapes of sperm and oocytes are very different from each other. Shortly after fertilization, the sperm-derived paternal chromatin and the oocyte-derived maternal chromatin show epigenetic modifications totally different from each other. During preimplantation development, the epigenetic asymmetry between the parental chromatins is gradually resolved. This resetting process, called epigenetic reprogramming, is believed to be important to prepare a white canvas to establish new epigenetic information in a cell type-specific manner during cellular differentiation at post-implantation.
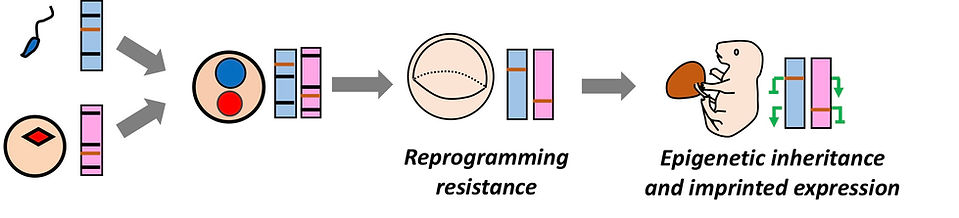
Reprogramming resistance & Genomic imprinting
While the epigenetic landscapes of gametes are genome-widely reprogrammed during preimplantation development, certain genomic loci escape the reprogramming wave. Such reprogramming resistance results in transmission of parental epigenetic information into the next generation. When the epigenetic modifications at reprogramming resistant loci are parent-of-origin specific, it causes imprinted expression. The representatives of these loci are imprinting control regions that are marked by oocyte- or sperm-specific DNA methylation.
Maternal inheritance of histone modification
Since the first identification of imprinted genes in mammals in 1991, DNA methylation had been the only known mechanism governing genomic imprinting. Nevertheless, a few studies have demonstrated that several paternally-expressed imprinted genes (PEGs) maintain their imprinted expression even in the lack of oocyte DNA methylation, suggesting the existence of a DNA methylation-independent imprinting mechanism.
In 2017, we discovered that oocyte H3K27me3 is transmitted into embryos and the oocyte-derived H3K27me3 controls all of the PEGs previously reported to be independent of germline DNA methylation (Inoue et al., 2017 Nature). Maternal H3K27me3-mediated imprinting is nowadays called “noncanonical imprinting”, while DNA methylation-mediated imprinting is called “canonical imprinting”. These PEGs include Xist, an X-linked long non-coding RNA critical for X chromosome inactivation (XCI). Noncanonical imprinting at Xist accounts for the long-sought mechanism of paternal X-selective inactivation during preimplantation and placental development, the process known as imprinted XCI (Inoue et al., 2017 Genes Dev; Inoue et al., 2018 Genes Dev). While canonical imprinting is maintained in both embryonic and extra-embryonic cell lineages, noncanonical imprinting is maintained only in extra-embryonic cells, such as placentae and yolk sacs.
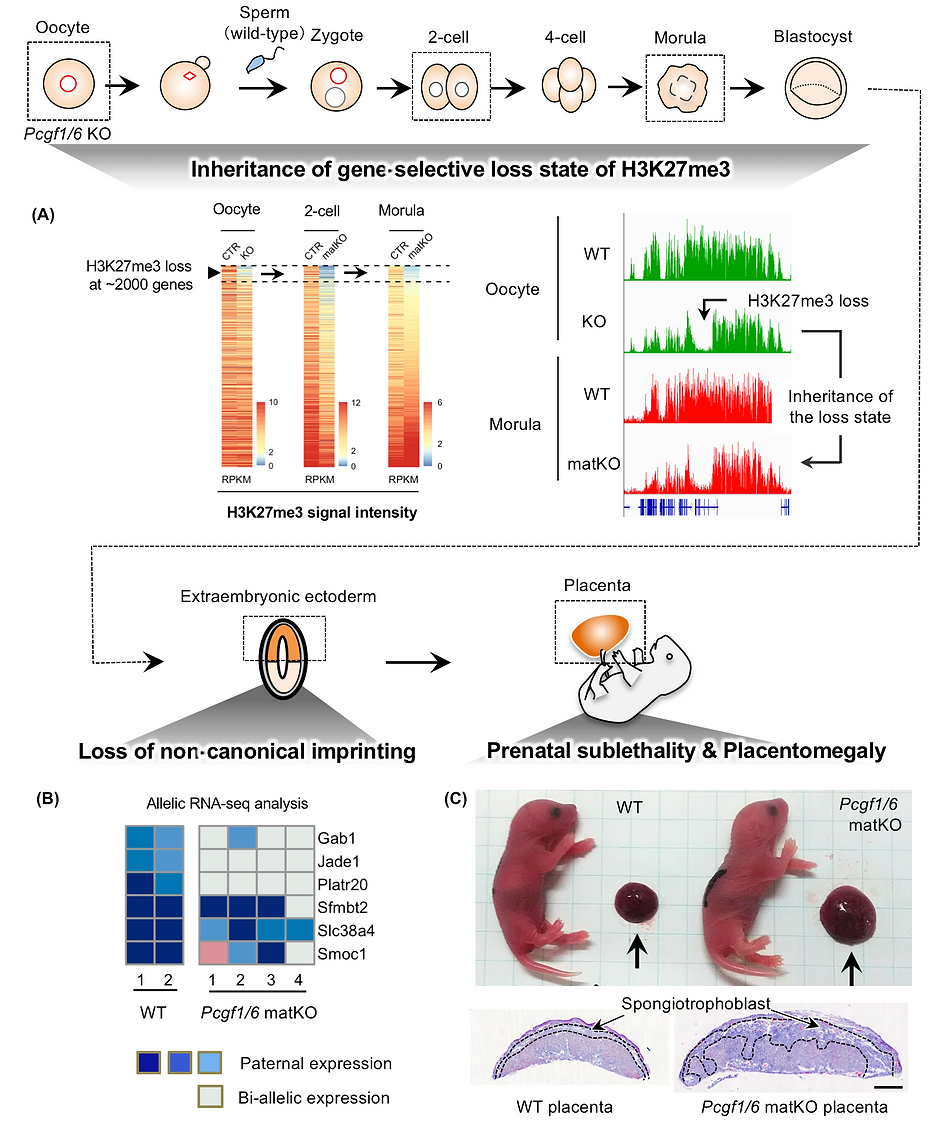
A loss-of-noncanonical imprinting (LOI) mouse model shows developmental retardation at post-implantation, prenatal sublethality, and placental enlargement (Mei et al., 2021 Nat Genet, Matoba, Kozuka et al., 2022 Genes Dev). Restoration of imprinted gene expression of Xist and several autosomal imprinted genes (chromosome 2 microRNA cluster and Slc38a4) rescues embryonic lethality and placental abnormalities, respectively, in LOI embryos, proving that noncanonical imprinting sustains mouse development and restrains placental overgrowth (Matoba, Kozuka et al., 2022 Genes Dev).
Future investigations are awaited to understand how noncanonical imprinting is regulated during development, what the functions are, why it is specific to the extraembryonic tissues, and why it has to be histone, but not DNA methylation.

Representative research in the lab-1
(H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Mei et al., 2021 Nat Genet)
To investigate how Polycomb repressive complex 2 (PRC2)-mediated H3K27me3 is established during oogenesis and how it is passed on to the next generation, we studied the role of PRC1-mediated histone H2A lysine 119 mono-ubiquitination (H2AK119ub1).
Using low-input CUT&RUN, we investigated the dynamics of H2AK119ub1 during oogenesis and early embryogenesis, and found that broad H2AK119ub1 domains are established, along with H3K27me3, during oocyte growth. On the other hand, these modifications differently behave after fertilization, with H2AK119ub1 being established prior to H3K27me3 at developmental genes that are typical PRC targets. As far as we know, this is the first report to show that H2AK119ub1 guides H3K27me3 in vivo.
To determine whether H2AK119ub1 is required for establishment of H3K27me3 during oogenesis, we generated oocytes lacking polycomb group ring finger 1 (PCGF1) and PCGF6, which are essential components of variant PRC1. Pcgf1/6 KO oocytes showed that H2AK119ub1 is globally reduced and H3K27me3 is not established at a subset of genes (about 2,000 genes) (Below Fig. A). Profiling of H3K27me3 distribution in maternal KO (matKO) embryos, obtained by fertilization of Pcgf1/6 KO oocytes with WT sperm, revealed that H3K27me3-lost genes continued to be devoid of H3K27me3 after fertilization, even in the presence of active PRC2 (Fig. A). After implantation, Pcgf1/6 maternal KO embryos showed bi-alleic expression of non-canonical imprinted genes (Fig. B), partial embryonic lethality, and placental enlargement in the survivors (Fig. C). Thus, if H3K27me3 is failed to be established in oocytes, it can no longer be reversed even if H3K27me3-modifying enzymes are present.
The phenotype of placental enlargement can be interpreted as the mother suppressing the size of the placenta via histone modifications in the oocytes. This means that non-canonical imprinting supports the conflict hypothesis (one of the evolutionary theories of genomic imprinting). This paper enriched interesting aspects of Polycomb molecular biology and genomic imprinting.
Representative research in the lab-2
(H3K27 di-methylation dynamics reveal stepwise establishment of facultative heterochromatin in early mouse embryos. Matsuwaka et al., 2025 Nat Cell Biol)
Totipotent zygotes have relaxed chromatin with little typical heterochromatin. Before implantation, H3K27me3 is deposited at >2,000 developmental gene promoters & inactivating paternal X-chr (Xp), forming facultative heterochromatin. How does the H3K27me3-mediated heterochromatin form de novo?
Of course, H3K27me3 is deposited by PRC2, the sole enzyme of H3K27 methylation. We know that PRC2 starts to deposit H3K27me3 from the blastocyst stage. But what is happening before that? How is PRC2 activated and recruited to chromatin?
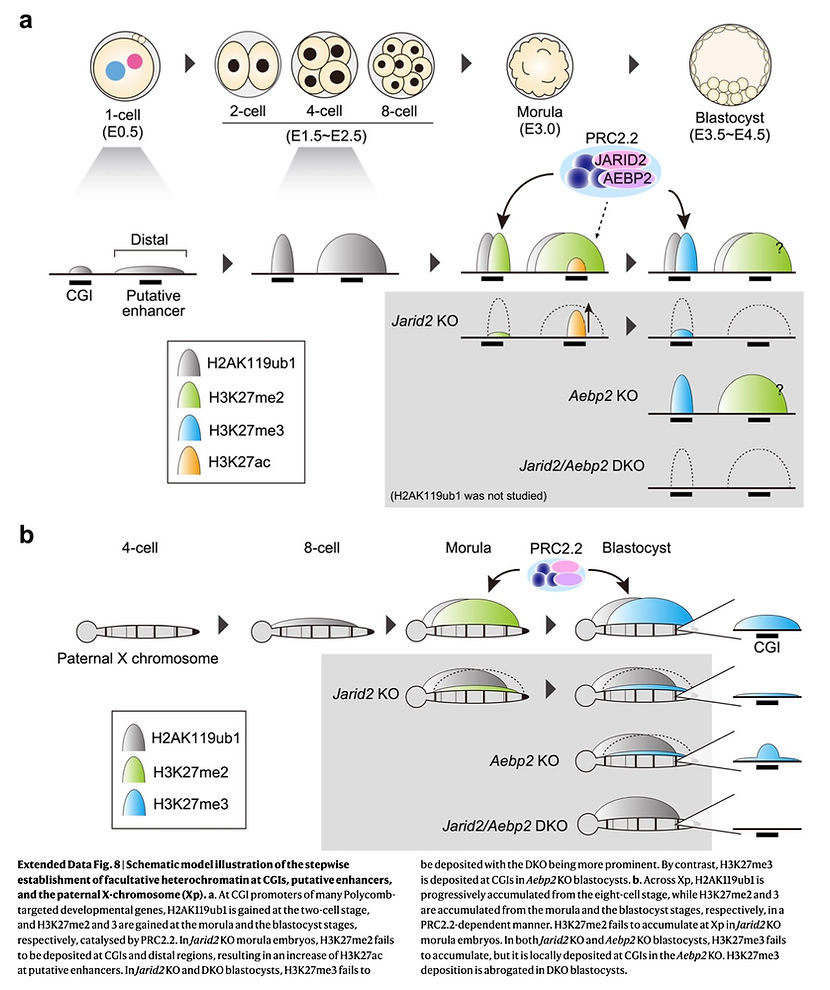
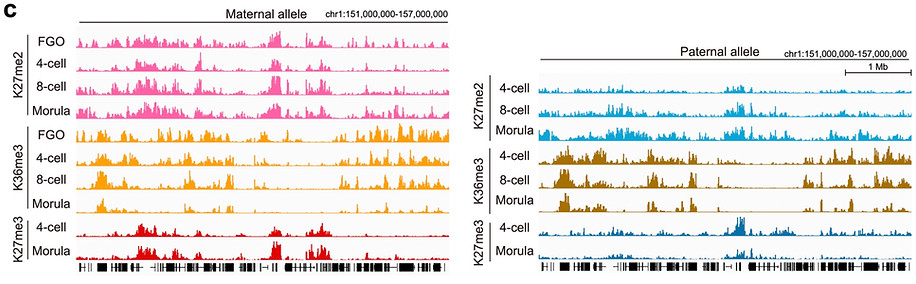
To address these, we decided to profile the dynamics of H3K27me2, a precursor of H3K27me3, in oocytes and preimplantation embryos, which allowed us to delineate the establishing kinetics. Integrative analysis with H3K27me3 and PRC1-deposited H2AK119ub1 revealed that facultative heterochromatin is formed in a stepwise manner –H2Aub1 first (2C at dev gene promoters and 8C at Xp), H3K27me2 second (morula) and then H3K27me3 (blastocyst).
Then, how does PRC2 get to its target chromatin in morula embryos? We found that JARID2 –a PRC2.2-specific subunit possessing H2AK119ub1-binding ability– is highly expressed in morula embryos, and binds to dev genes & Xp together with SUZ12 (a core subunit of PRC2) in morulae.
By making Jarid2 KO morulae, we showed that JARID2 is required for SUZ12 chromatin binding and H3K27me2 deposition. Furthermore, loss of H3K27me2 over intergenic regions led to an increase of H3K27ac at putative enhancers, suggesting that H3K27me2 may prevent promiscuous enhancer activation in embryos.In Jarid2 KO blastocysts, H3K27me3 was poorly deposited at dev genes & Xp. Moreover, parallel analysis of Aebp2 (another PRC2.2-specific subunit) KO and Jarid2/Aebp2 DKO blastocysts revealed the unique and synergistic roles of AEBP2 for H3K27me3 deposition at dev genes and across Xp. These data indicate that facultative heterochromatin is slowly formed by the H2AK119ub1-reading PRC2.2 complex in a stepwise manner.
Unique research tools
In addition to general molecular biology tools, we have unique research tools as listed below. The ability to integrate these allows us to conduct high-originality researches.
1. Epigenome analysis technologies applicable to a small number of cells
Carrier DNA-assisted ChIP-seq (CATCH-seq) (Matsuwaka et al., 2025 Nat Cell Biol)
Low-input DNaseI-seq (Lu et al., 2016 Cell, Inoue et al., 2017 Nature)
Low-input CUT&RUN (Hayashi and Inoue, 2023 Method Mol Biol)
Low-input RRBS(Shen et al., 2014 Cell Stem Cell, Inoue et al., 2015 Cell Rep)
etc.
2. Micromanipulation & Developmental engineering
mRNA/siRNA injection, pronuclear isolation, pronuclear transfer, chromosome transfer, somatic cell nuclear transfer, in vitro fertilization (IVF), in vitro maturation (IVM), in vitro growth (IVG), intracytoplasmic sperm injection (ICSI), embryo transplantation, dissection of E6.5 epiblast/extra-embryonic ectoderm/visceral endoderm, etc.
3. Maternal protein depletion system
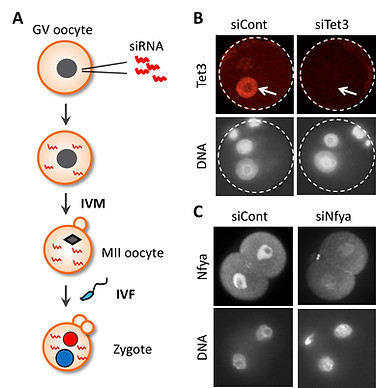
siRNA injection into GV oocytes followed by IVM-IVF
Since most of biological events occurring in zygotes and early preimplantation embryos are regulated by maternally-stored proteins, it is essential to deplete them to understand their mechanisms and functions. One way is to generate an oocyte-specific KO model, which is time-consuming and thereby extremely low throughput. An alternative is RNA interference-mediated knockdown of interested genes. Although siRNA injection into MII-stage oocytes is commonly used due to its relative easiness, it often fails to deplete maternal proteins because these proteins have already been accumulated in MII oocytes. To achieve efficient knockdown of maternal proteins, we inject siRNA into GV-stage oocytes and then let the oocytes reach the MII-stage using in vitro maturation (IVM). This allows a 20-42 hrs time window for injected siRNA to act before fertilization. The usefulness of this approach has been proved by our past works in which maternally-stored proteins, Tet3 and Nyfa, were efficiently depleted (Inoue et al., 2012 Cell Res; Lu et al., 2016 Cell). The detailed protocol will be distributed upon request.
siRNA injection into growing oocytes followed by IVG-IVM-IVF
Nevertheless, it sometimes happens that the above approach fails to knockdown targeted maternal proteins when the proteins are highly stable. To overcome this issue, we inject siRNA into growing oocytes followed by in vitro growth (Inoue et al, 2014 Protocol Ex). In this system, secondary follicles are collected from ovaries of 12 day-old mice, oocytes within the follicles are injected with siRNA, and then the follicles are cultured in vitro for 12 days until the oocytes reach the fully-grown GV-stage. Then, the oocytes are matured in vitro (IVM) and fertilized in vitro (IVF) to obtain zygotes. This system provides a long time window (~2 weeks) for injected siRNA to act. The injected siRNA can act for such a long period because the oocyte is a non-dividing cell and siRNA is not diluted by cell division. By using this method, we have succeeded to knockdown NPM2 and HIRA in mouse oocytes (Inoue and Aoki, 2010 FASEB J; Inoue and Zhang, 2014 NSMB).